Neural Network Basics
Derive the gradients of the sigmoid function.
So,the sigmoid function is:
$$sigmiod(x) = \sigma (x)$$
$$ \sigma ‘(x) = - \frac{1}{(1+e^{-x})^2} \cdot -e^{-x} $$
$$ = \frac{1}{1+e^{-x}} \cdot \frac{e^{-x}}{1+e^{-x}} $$
$$ = \frac{1}{1+e^{-x}} \cdot (1 - \frac{1}{1+e^{-x}}) $$
$$ = \sigma(x) \cdot (1-\sigma(x)) $$
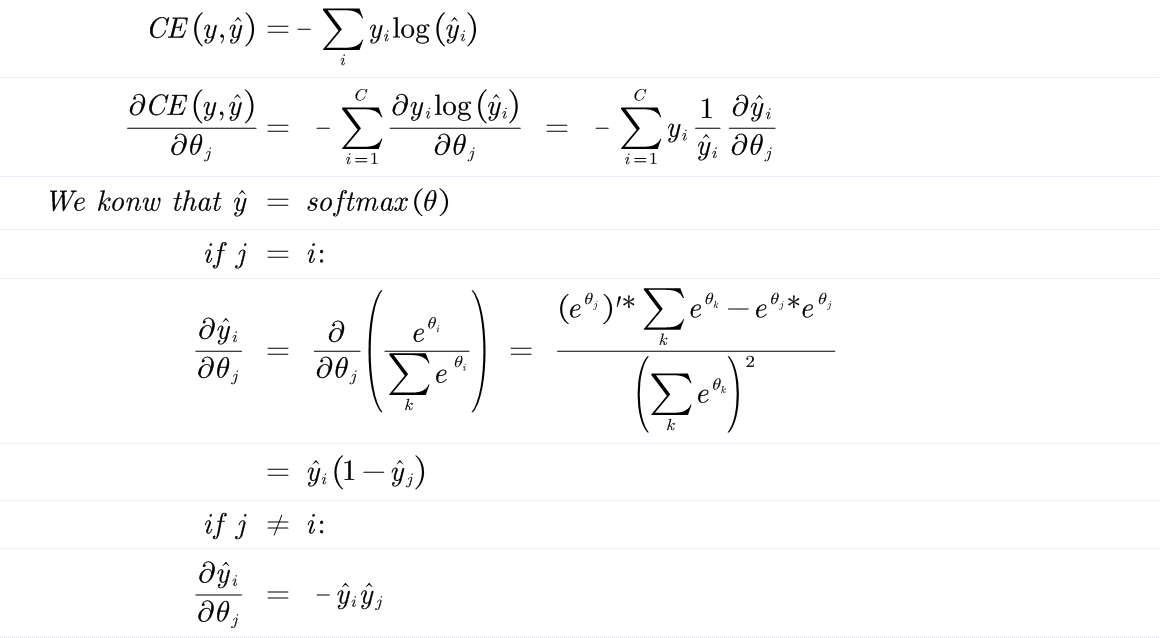
Derive the gradient of the cross entropy function with the softmax fuction.
$$ CE(y,\hat{y}) = - \sum_i y_i log(\hat{y_i}) $$
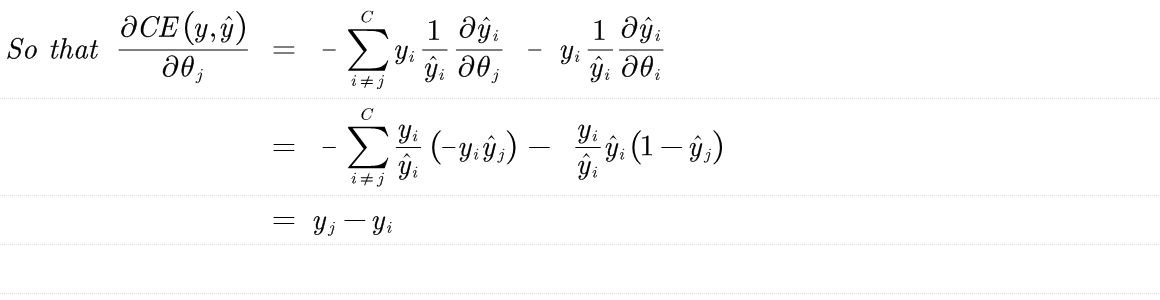
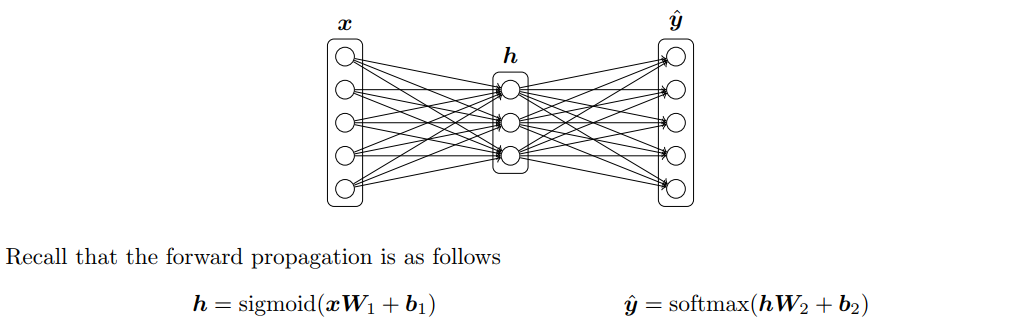
Derive the gradients with respect to the inputs x to an one-hidden-layer neural network.
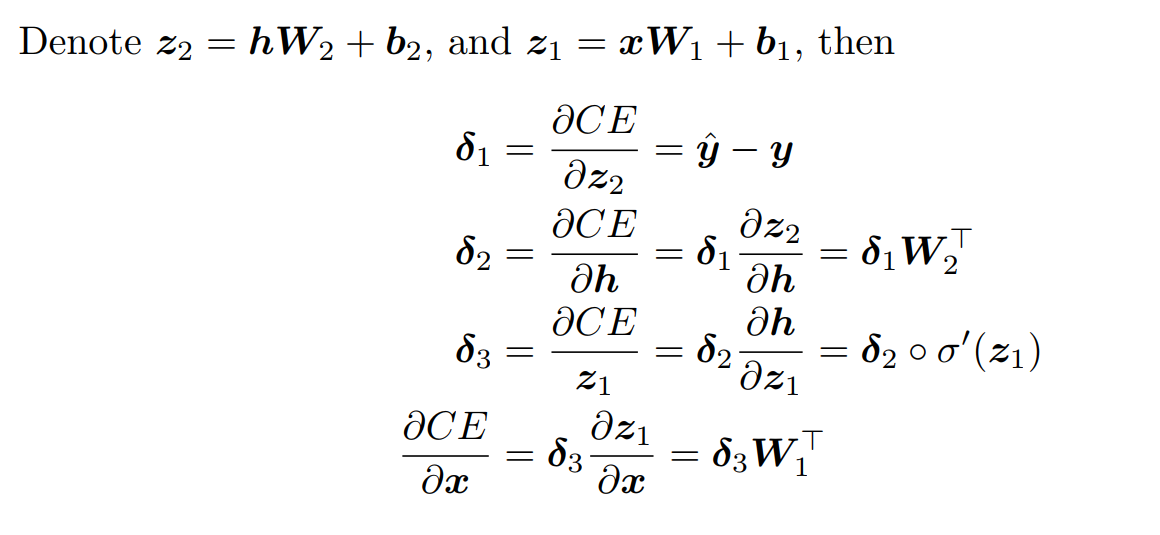
Calculate the value of parameters in this neural network:
assuming the input is $D_x$-dimensional,the output is $D_y$-dimensional, and there are H hidden units?
Similar to the neural network in c:
The anwser is:
(Dx+1)H+(H+1)Dy
Fill in the implementation for the sigmoid activation function and its gradient
From the mathematical point of view,The sigmoid function should do it:
|
|
And then,my improvement of the gradient for the sigmoid function :
|
|
Now you can test iy by calling:
|
|
Running basic tests...
[[ 0.73105858 0.88079708]
[ 0.26894142 0.11920292]]
[[ 0.19661193 0.10499359]
[ 0.19661193 0.10499359]]
Now,we use the implemented sigmoid function to create the graph to understand the behavior of this function:
|
|
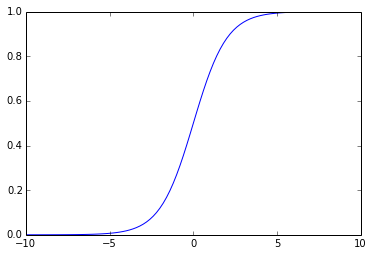
From the above graph,we can observe that the sigmoid function produces the curve which will be in shape “S”,and returns the output value which falls in the range of 0 to 1.
The below are the properties of the Sigmoid function:
- The high value will have the high score but not the higher score.
- Used for binary classification in logistic regression model.
- The probabilities sum need not be 1.
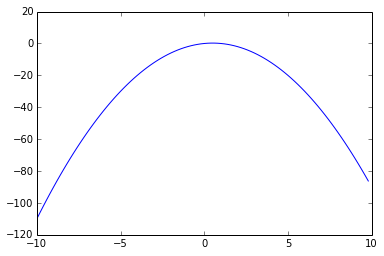
The below is the graph of the gradient of the sigmoid function:
|
|
To make debugging easier, we will now implement a gradient checker. Fill in the implementation for gradcheck naive in q2 gradcheck.py.
Gradient checks
In theory,performing a gradient check is as simple as comparing the analytic gradient and the numerical gradient.We use the centered difference formula of the form:
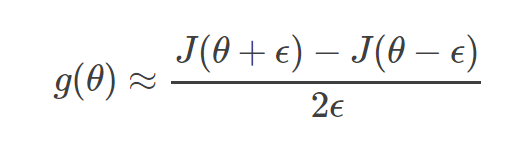
where ϵ is a very small number,in practice approximately le-5 or so.
Use relative for the comparison.
It is always more appropriate to consider the relative error.
- relative error > le-2 usually means the gradient is probably wrong.
- le-7 and less you should be happy.
That means,to implement the gradient check,we would need:
|
|
Then generate trial function that are used by multiple tests:
|
|
Running sanity checks...
Gradient check passed!
Gradient check passed!
Gradient check passed!
|
|
Running your sanity checks...
The original function is: ln(x)
Gradient check passed!
The original function is: sin(x)-cos(x)
Gradient check passed!
Gradient check passed!
Gradient check passed!
The original function is: exp(x)
Gradient check passed!
Gradient check passed!
Gradient check passed!
implement the forward and backward passes for a neural network with one sigmoid hidden layer.
Let’s consider a simple logistic regression classifier.
The forward pass computes the output given the input for inference.In forward composes the computation of each layer to computation of each layer to compute the “function” represented by the model.
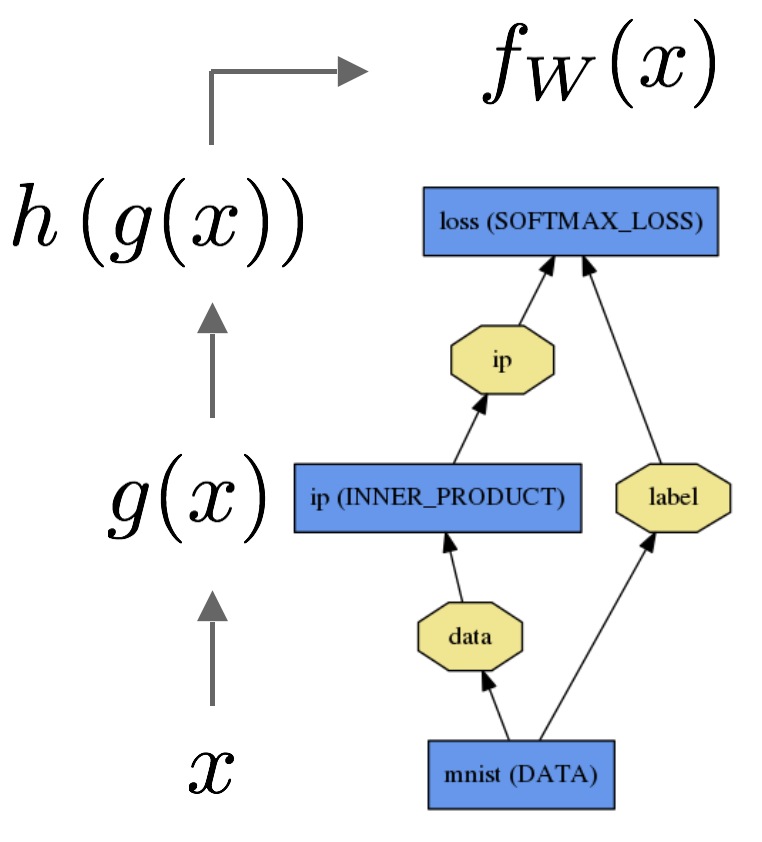
The backward pass computes the gradient given the loss for learning.In backward composes the gradient of each layer to compute the gradient of the whole model by automatic differentiation.
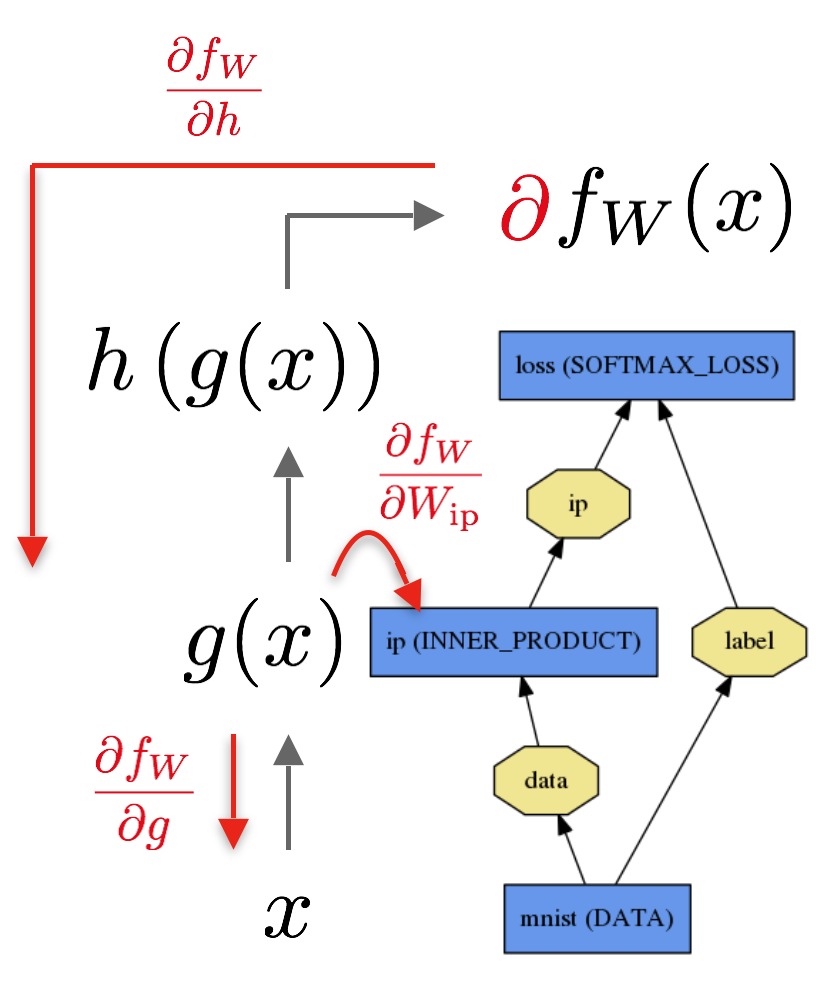
The below is my implementation:
|
|
Then generate trial function that is used by test:
|
|
Running sanity check...
Gradient check passed!